Faster Ordinary Differential Equations (ODEs) solvers and Sensitivity Analysis of Parameters: Introducing SUNDIALS support in MATLAB
Late last year I introduced the new solution framework for solving Ordinary Differential Equations (ODEs) that made its debut in MATLAB R2023b. I demonstrated how it allowed users to do all kinds of things much more easily than before but stressed that the R2023b release was mostly about the new interface. I told you "There are no new solvers or any fundamentally new functionality....yet!". A few people picked up on the 'yet', correctly guessing that there would be new solvers and functionality soon. In R2024a, we've made a start on this by adding support for some of the SUNDIALS solvers.
What is SUNDIALS?
SUNDIALS is a SUite of Nonlinear and DIfferential/ALgebraic equation Solvers, an award-winning set of open source ODE solvers from Lawrence Livermore National Labs. Functionality from SUNDIALS has been available via Simbiology for a while and now we've brought it to all MATLAB users via the new ODE solver interface. There are several solvers in the SUNDIALS suite and we've added support for three of them via new values of the Solver property of the ode class: "cvodesstiff", "cvodesnonstiff" and "idas".
- "cvodesnonstiff" Variable-step, variable-order (VSVO) solver using Adams-Moulton formulas, with the order varying between 1 and 12. See CVODE and CVODES.
- "cvodesstiff" Variable-step, variable-order (VSVO) solver using Backward Differentiation Formulas (BDFs) in fixed-leading coefficient form, with order varying between 1 and 5. See CVODE and CVODES.
- "idas" Variable-order, variable-coefficient solver using Backward Differentiation Formulas (BDFs) in fixed-leading coefficient form, with order varying between 1 and 5. See IDA and IDAS.
An example: How to switch to a SUNDIALS solver in MATLAB
Let's begin by solving the classic predator-prey equations
with parameters and initial conditions
Setting up and solving the problem using the ode class looks like this
d = ode(ODEFcn=@(t,y,p) [p(1)*y(1) - p(2)*y(1)*y(2); ...
p(3)*y(1)*y(2) - p(4)*y(2)]);
% Set initial conditions and parameters
d.InitialTime = 0;
d.InitialValue = [1;1];
d.Parameters = [.4 .4 .09 .1];
% Set the relative tolerance
d.RelativeTolerance = 1e-6;
At this point, I could just get on and solve the system of ODEs with the solve function and MATLAB would attempt to choose a suitable solver for me and in R2024a it happens to choose ode45 for this problem. This could change in future versions though so let's explicitly choose ode45 so we know exactly what we'll be using.
% Select the ode45 solver
d.Solver = "ode45";
% Solve over a short time span
sol = solve(d,0,100);
plot(sol.Time,sol.Solution,LineWidth=2)
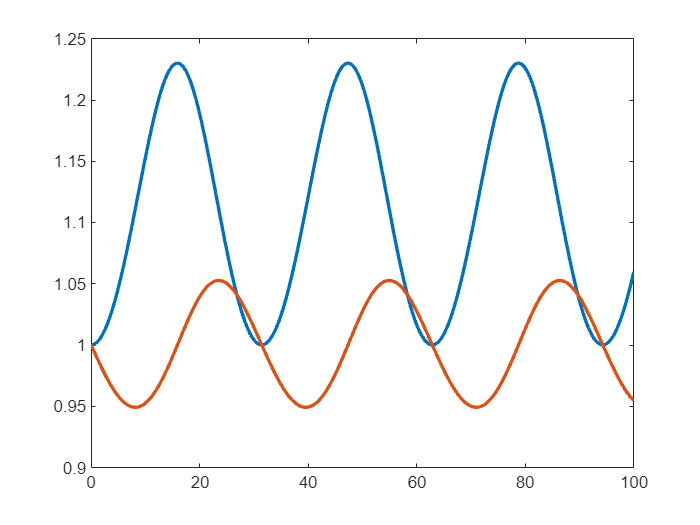
To switch to using the non stiff version of SUNDIAL's CVODES solver, I just need to do
% select the cvodesnonstiff solver
d.Solver = "cvodesnonstiff";
and then solve as before
% Solve over a short time span
sol = solve(d,0,100);
plot(sol.Time,sol.Solution,LineWidth=2)
Not much appears to have changed! So what are the benefits of using the SUNDIALS solvers?
SUNDIALS solvers can lead to improved performance
The first potential benefit of the new SUNDIALS solvers is performance. I'm going to switch back to ode45, solve over a much larger time span and use the timeit function to get solution time
d.Solver = "ode45";
timeOde45Fcn = @() solve(d,0,15000);
ode45Time = timeit(timeOde45Fcn)
Now let's solve the exact same problem with SUNDIALS' CVODES non stiff solver.
d.Solver = "cvodesnonstiff";
timeCvodesFcn = @() solve(d,0,15000);
cvodesTime = timeit(timeCvodesFcn)
Let's see what the speed-up is
fprintf("The SUNDIALS solver cvodesnonstiff is %f times faster than ode45 for this problem\n",ode45Time/cvodesTime)
I was pretty pleased with this! Internal presentations about the new solver suggested that the speed-up would be ~2x or so for most benchmarks and I've gotten way more than that here. The reasons for the speed-up of cvodesnonstiff compared to ode45 are primarily that it requires fewer steps and its slightly faster per step. The SUNDIALS suite is written in C and we've also performed some JIT magic to reduce the overhead of calling a MATLAB function from a C++ function.
We can get some insight into this by looking at the solutions. Over the time span [0,15000], ode45 takes 33409 steps for the tolerance I have set
d.Solver = "ode45";
ode45Sol = solve(d,0,15000)
while cvodesnonstiff takes 11503 steps
d.Solver = "cvodesnonstiff";
cvodesSol = solve(d,0,15000)
That's 2.9x fewer steps than ode45 but there's more than a 2.9x speed-up so the speed-difference is clearly not just because the new solver requires fewer steps.
I've been comparing against ode45 because that's the algorithm I always reach for first. However, cvodesnonstiff is most similar to ode113 as far as the algorithm under the hood is concerned and it turns out that ode113 requires about the same number of steps as cvodesnonstiff.
d.Solver = "ode113";
ode45Sol = solve(d,0,15000)
Very close in terms of number of steps but it turns out that ode113 is even slower than ode45 for this problem
d.Solver = "ode113";
timeOde113Fcn = @() solve(d,0,15000);
timeOde113 = timeit(timeOde113Fcn)
fprintf("The SUNDIALS solver cvodesnonstiff is %f times faster than ode113 for this problem\n",timeOde113/cvodesTime)
When I spoke to development about these speed differences they told me that I happened to choose a problem that really allows the new SUNDIALS integration to shine! For example, I chose an integration time that was large enough to dwarf the startup cost. For various reasons, the overhead for getting from ode object into the SUNDIALS solver is generally bigger than getting into ode45 and friends. Once we are in SUNDIALS, however, things can really fly for small problems like this.
For bigger, stiff problems, we find less of a performance difference between the MATLAB and SUNDIALS solvers. The way this was explained to me is to consider what the solver actually does. It evaluates a function handle and then does some vector-vector or matrix-vector operations/solves/etc. When the problems get big, the linear algebra starts to dominate and the time per step will be similar for both solvers. At this point, the winning solver is the one that happens to have fewer steps or which one that manages to trigger fewer matrix factorizations etc, which again, is problem dependent.
So, the SUNDIALS can be a lot faster than ode45 and friends...and sometimes its not. The exact speed difference between solvers depends heavily on the problem and I encourage you to investigate on your own problems and let me know what you find.
SUNDIALS solvers allow sensitivity analysis of parameters in systems of ODEs
Having a faster ODE solver in MATLAB is great but there is another reason to love the SUNDIALS integration: Sensitivity analysis of parameters. This is functionality that Simbiology users have had for a while but now some of it is in MATLAB itself. Sensitivity analysis examines how changes in parameter values can affect the solution of the ODE system.
The MATLAB documentation demonstrates this by looking at a Sensitivity analysis of a SIR epidemic model so I went looking for a different model to play with. I soon found the CARRGO model [1] via a paper about methods for assessing sensitivity in biological models [2]. It models the interaction of cancer cells and CAR T-cells which are used to destroy them and is a variation of Lotka-Volterra predator-prey equations. The system of ODEs is
x is the density of cancer cells andy is the density of CAR T-cells. In MATLAB, I'll refer to the parameters as a vector p and since there are 5 parameters here, we'll have a 5 element vector. The meaning of each parameter and the number I'll give it in MATLAB is as follows
- = p(1): The killing rate of the CAR T-cells.
- = p(2): Net rate of proliferation of CAR T-cells
- θ= p(3): Death rate of CAR T-cells
- ρ= p(4): Net growth rate of cancer cells
- γ= p(5): Cancer cell carrying capacity
Sensitivity analysis will give us the derivatives of x(density of cancer cells) and y(density of CAR T-cells) with respect to these five parameters.
Setting the equations up in MATLAB:
F = ode();
F.ODEFcn = @(t,x,p) [ p(4) * x(1) * (1-x(1) / p(5))-p(1) * x(1) * x(2); p(2)* x(1) * x(2)-p(3) * x(2)];
Sahoo et al [1] fitted this model to empirical data (Using MATLAB and fmincon so the paper tells us) and found the following parameters and initial values
F.InitialValue = [1.25*10^4 , 6.25*10^2];
F.Parameters = [6*10^-9, 3*10^-11, 1*10^-6, 6*10^-2, 1*10^9];
Set the solver to cvodesnonstiff and we can get the solution to the system of equations as usual
F.Solver = "cvodesnonstiff";
sol = solve(F,0,1000)
Next, I plot these solutions
tiledlayout(2,1)
nexttile
plot(sol.Time,sol.Solution(1,:),LineWidth=2)
title("x : density of cancer cells")
xlabel("days")
ylim([-0.5*10^8 11*10^8])
nexttile
plot(sol.Time,sol.Solution(2,:),LineWidth=2)
title("y : density of CAR T-cells")
xlabel("days")
ylim([-0.5*10^6 16*10^6])
sgtitle("Solutions of the CARRGO system of ODEs")
Computing the parameter sensitivities
Computing the sensitivities of the system of equations to all 5 parameters is very easy. We simply add the following to the ODE object.
F.Sensitivity = odeSensitivity();
and run the solve command as before.
sol = solve(F,0,1000)
Note that this time, our ODEResults has a Sensitivity field alongside the Solution. There are 2 x 5 sets of Sensitivity results corresponding to the partial derivatives of either of our 2 variables with respect to the 5 parameters. If I wanted to only consider a few indices, I would change ParameterIndices accordingly.
You're going to need to squeeze!
Let's get the result for the first variable, x, and the first parameter, , i.e.
dXdKappa1 = sol.Sensitivity(1,1,:);
sensitivitySize = size(dXdKappa1)
The problem with this is that it is a 1 x 1 x 225 array so plotting it directly won't work!
plot(sol.Time,dXdKappa1)
You have to first squeeze it to get rid of the redundant singleton dimension. You'll find that there is a lot of squeezing to be done when dealing with sensitivities!
figure
plot(sol.Time,squeeze(dXdKappa1),LineWidth=2)
ylabel("$\frac{\partial x}{\partial\kappa_1}$",Interpreter="latex")
xlabel("days");
title("Sensitivity of Cancer Cells in the CARGGO Model to the $\kappa_1$ parameter",Interpreter="latex")
The result is the partial derivative of x with respect to the parameter
Normalizing and comparing the sensitivity functions
The epidemic example in the MATLAB documentation for Sensitivity Analysis tells us that sensitivity functions are often normalized such that the result describes the approximate percentage change in each solution component due to a small change in the parameter. This helps us compare the effect of each of the parameters on the overall model. Refer to the doc page or Hearne [3] for the mathematics. The code looks like this for the first variable,that represents cancer.
p = F.Parameters;
normalisedSensitivityKappa1 = squeeze(sol.Sensitivity(1,1,:))'.*(p(1)./sol.Solution(1,:));
normalisedSensitivityKappa2 = squeeze(sol.Sensitivity(1,2,:))'.*(p(2)./sol.Solution(1,:));
normalisedSensitivityTheta = squeeze(sol.Sensitivity(1,3,:))'.*(p(3)./sol.Solution(1,:));
normalisedSensitivityRho = squeeze(sol.Sensitivity(1,4,:))'.*(p(4)./sol.Solution(1,:));
normalisedSensitivityGamma = squeeze(sol.Sensitivity(1,5,:))'.*(p(5)./sol.Solution(1,:));
Plot each one of these against time
figure
t = tiledlayout(3,2);
title(t,"Normalised sensitivty functions for Cancer in the CARRGO model",Interpreter="latex")
xlabel(t,"Time (days)",Interpreter="latex")
ylabel(t,"\% Change in Eqn",Interpreter="latex")
nexttile
plot(sol.Time,normalisedSensitivityKappa1,LineWidth=2)
title("$\kappa_1$",Interpreter="latex")
ylim([-1.5 0.5])
nexttile
plot(sol.Time,normalisedSensitivityKappa2,LineWidth=2)
title("$\kappa_2$",Interpreter="latex")
ylim([-30 0.5])
nexttile
plot(sol.Time,normalisedSensitivityTheta,LineWidth=2)
title("$\theta$",Interpreter="latex")
ylim([-0.005 0.01]);
nexttile
plot(sol.Time,normalisedSensitivityRho,LineWidth=2)
title("$\rho$",Interpreter="latex")
ylim([-6 10])
nexttile
plot(sol.Time,normalisedSensitivityGamma,LineWidth=2)
title("$\gamma$",Interpreter="latex")
ylim([-21 5])
I am not a biologist but my naive expectation was that , the killing rate of the CAR T-cells would have a strong effect on the model. However, the above plots suggest that this isn't the case. For almost half of the simulation, this rate has no effect at all and when it does kick in, it's much weaker than the other parameters. $ \kappa_2~ $, the net rate of proliferation, is much more important and that too only starts to have an effect roughly half way through the simulation. The parameter θ seems to have very little affect at all.
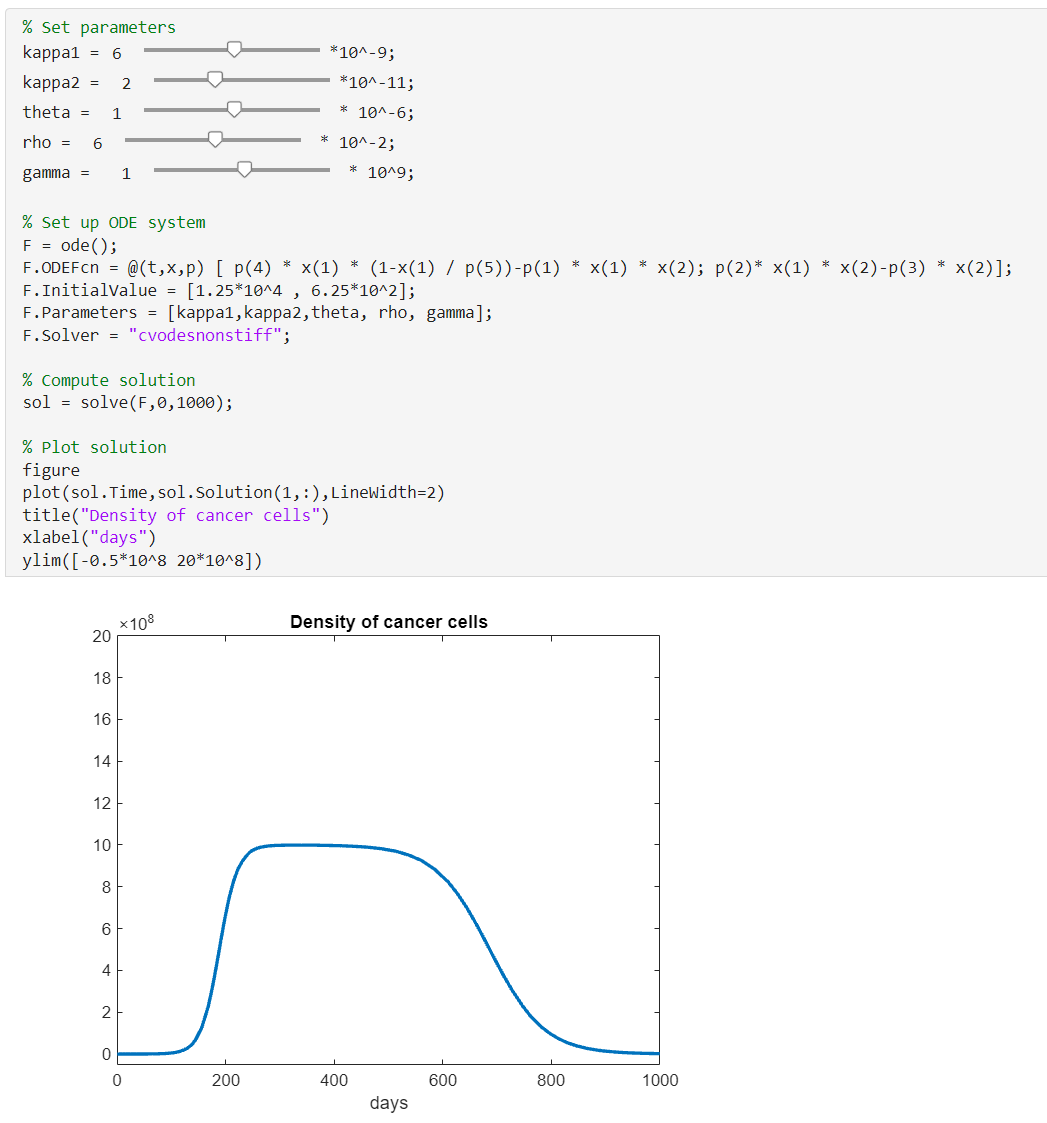
To explicitly see these conclusions, and any others you might make from the above plots, I used live controls to create a mini application based on this model that allows the user to vary the all of the parameters around the values we've been working with. Sure enough, θ doesn't make much of a difference and the observations around $ \kappa_1 $ and $ \kappa_2 $ play out too.
Click on the image below to open this model up in MATLAB Online to have a play yourself.
More advanced sensitivity analysis
The type of sensitivity analysis considered here is a “local, forward sensitivity analysis”. There's a lot more to sensitivity analysis than I've shown here and you may be interested in features more advanced than those added to the ode class in R2024a. If so, I suggest you take a look at the sensitivity analysis functionality in Simbiology which includes much more.
References
[1] Sahoo P, Yang X, Abler D, Maestrini D, Adhikarla V, Frankhouser D, Cho H, Machuca V, Wang D, Barish M, Gutova M, Branciamore S, Brown CE, Rockne RC. Mathematical deconvolution of CAR T-cell proliferation and exhaustion from real-time killing assay data. J R Soc Interface. 2020 Jan;17(162):20190734. doi: 10.1098/rsif.2019.0734. Epub 2020 Jan 15. PMID: 31937234; PMCID: PMC7014796.
[2] Mester R, Landeros A, Rackauckas C, Lange K. Differential methods for assessing sensitivity in biological models. PLoS Comput Biol. 2022 Jun 13;18(6):e1009598. doi: 10.1371/journal.pcbi.1009598. PMID: 35696417; PMCID: PMC9232177.
[3] Hearne, J. W. “Sensitivity Analysis of Parameter Combinations.” Applied Mathematical Modelling 9, no. 2 (April 1985): pp. 106–8. https://doi.org/10.1016/0307-904X(85)90121-0












コメント
コメントを残すには、ここ をクリックして MathWorks アカウントにサインインするか新しい MathWorks アカウントを作成します。