Cell segmentation
Blog reader Ramiro Massol asked for advice on segmenting his cell images, so I gave it a try. I'm not a microscopy expert, though, and I invite readers who have better suggestions than mine to add your comments below.
Let's take a look first to see what we have. I'm going to work with a cropped version of the original so that the images aren't too big for the layout of this blog.
Note: you can download the functions imcredit and imoverlay from MATLAB Central.
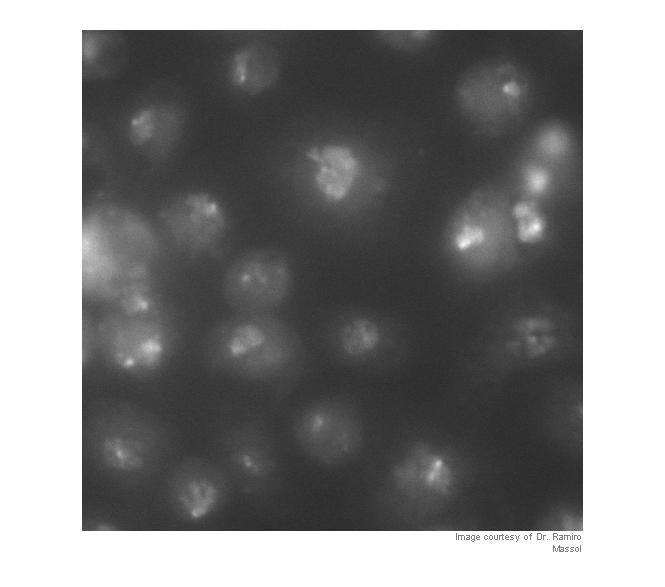
I = imread('https://blogs.mathworks.com/images/steve/60/nuclei.png'); I_cropped = I(400:900, 465:965); imshow(I_cropped) imcredit('Image courtesy of Dr. Ramiro Massol')
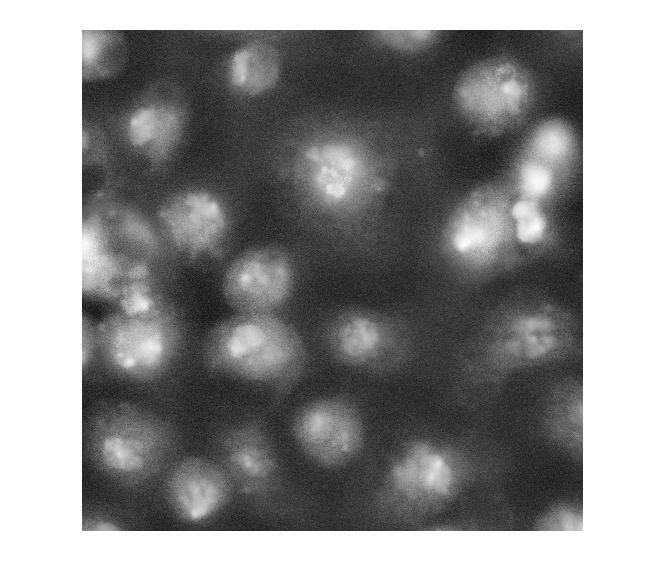
Strictly speaking, contrast adjustment isn't usually necessary for segmentation, but it can help the algorithm developer see and understand the image data better. This is a fairly low-contrast image, so I thought it might help. You can adjust the display contrast interactively with imtool, or you can use an automatic method such as adapthisteq. adapthisteq implements a technique called contrast-limited adaptive histogram equalization, or CLAHE. (I always thought "CLAHE" sounded like it must be some Klingon delicacy.)
I_eq = adapthisteq(I_cropped); imshow(I_eq)
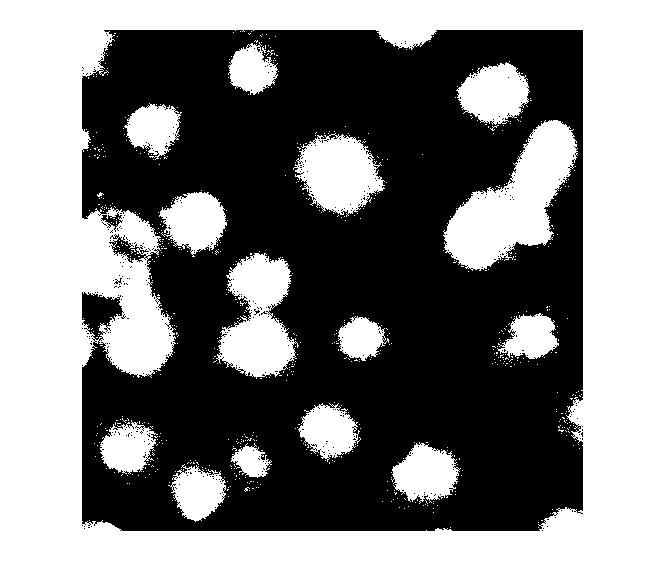
So what happens if we just apply a threshold now?
bw = im2bw(I_eq, graythresh(I_eq)); imshow(bw)
Let's clean that up and then overlay the perimeter on the original image.
bw2 = imfill(bw,'holes');
bw3 = imopen(bw2, ones(5,5));
bw4 = bwareaopen(bw3, 40);
bw4_perim = bwperim(bw4);
overlay1 = imoverlay(I_eq, bw4_perim, [.3 1 .3]);
imshow(overlay1)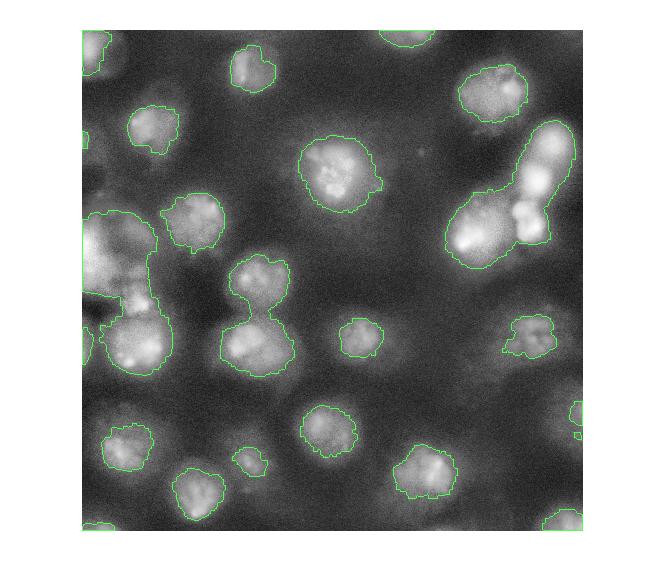
Now, I'm not familiar with these cell images, so I don't know exactly what I'm looking at. I assume some of these blobs need more help to be separated properly. One possible approach is called marker-based watershed segmentation. There's a demo of this idea on The MathWorks web site.
With this method, you have to find a way to "mark" at least a partial group of connected pixels inside each object to be segmented. You also have to mark the background.
Let's try to use the bright objects, which I assume are nuclei. The extended maxima operator can be used to identify groups of pixels that are significantly higher than their immediate surrounding.
mask_em = imextendedmax(I_eq, 30); imshow(mask_em)
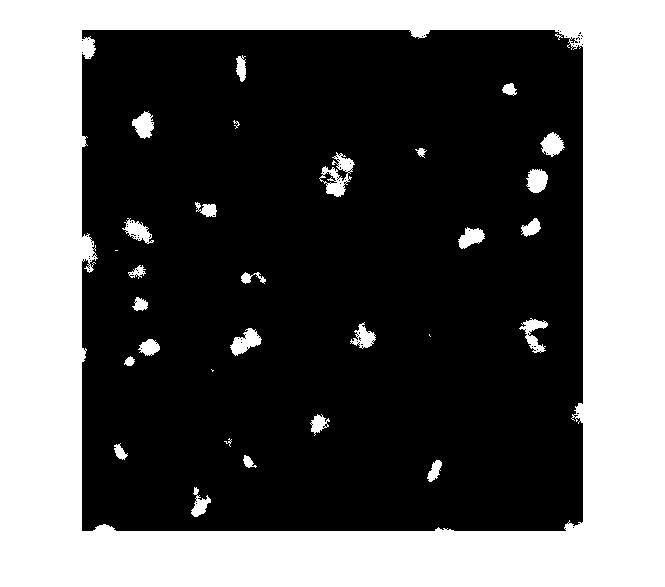
Let's clean that up and then overlay it.
mask_em = imclose(mask_em, ones(5,5));
mask_em = imfill(mask_em, 'holes');
mask_em = bwareaopen(mask_em, 40);
overlay2 = imoverlay(I_eq, bw4_perim | mask_em, [.3 1 .3]);
imshow(overlay2)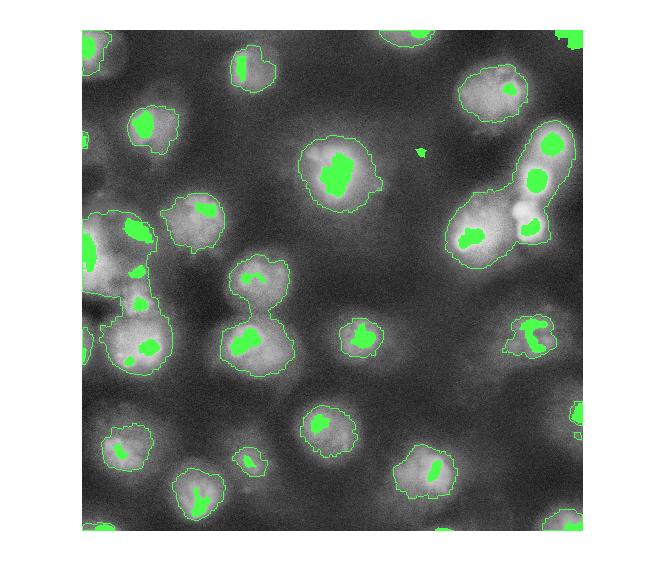
Next step: complement the image so that the peaks become valleys. We do this because we are about to apply the watershed transform, which identifies low points, not high points.
I_eq_c = imcomplement(I_eq);
Next: modify the image so that the background pixels and the extended maxima pixels are forced to be the only local minima in the image.
I_mod = imimposemin(I_eq_c, ~bw4 | mask_em);
Now compute the watershed transform.
L = watershed(I_mod); imshow(label2rgb(L))
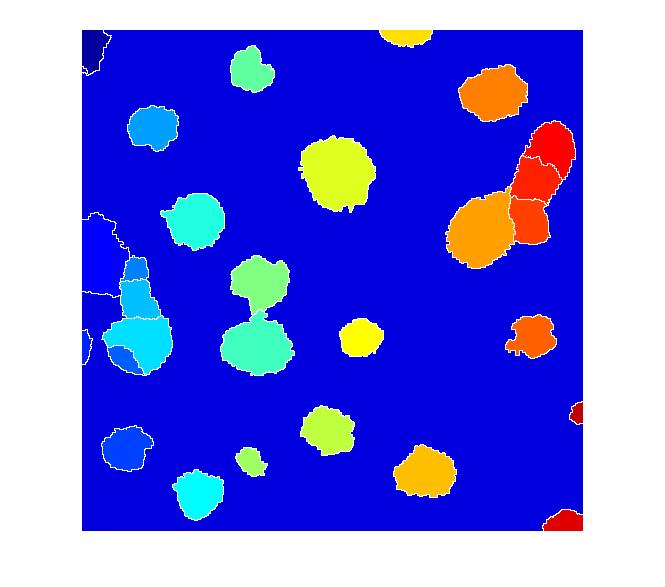
I don't know if this is a good segmentation result or not, but I hope some of the methods I've shown will give Dr. Massol some ideas to try.
Other readers ... if you have suggestions, I invite you to post your comments here.







댓글
댓글을 남기려면 링크 를 클릭하여 MathWorks 계정에 로그인하거나 계정을 새로 만드십시오.