This AI-augmented microscope uses deep learning to take on cancer
According to the American Cancer Society, cancer kills more than 8 million people each year. Early detection can boost survival rates. Researchers and clinicians are feverishly exploring avenues to provide early and accurate diagnoses, as well as more targeted treatments.
Blood screenings are used to detect many types of cancers, including liver, ovarian, colon and lung cancers. Current blood screening methods typically rely on affixing biochemical labels to cells or biomolecules. The labels adhere preferentially to cancerous cells, enabling instruments to detect and identify them. But these biochemicals can also damage the cells, leaving them unsuitable for further analyses.
The ideal approach would not require labels… but without a label, how can you get an accurate signal or any signal at all?
Combining microscopy and deep learning to diagnose cancer
To address this, researchers are developing new approaches using label-free techniques, where unaltered blood samples are analyzed based on their physical characteristics. Label-free cell analysis also faces limitations: The method can damage cells, due to the intense illumination required. Label-free cell analysis can also be highly inaccurate since it often relies on a single physical attribute. New research aims to improve accuracy while minimizing damage to cells.
In a recent issue of Nature Scientific Reports, a multidisciplinary team of UCLA researchers combined a new form of microscopy called photonic time-stretch imaging with deep learning. With this powerful new technique, they were able to capture 36 million video frames per second. (Compare that rate to human vision: We can theoretically process 1000 separate frames per second, but our brains blur images into motion at speeds high than 150 frames/second. Most television and films run at 24 frames/second.) They then used these images to detect cancerous cells.
.
The team at UCLA: A multidisciplinary approach
The team’s strategy was to devise a new and accurate label-free methodology by developing and integrating three key components into one system: quantitative phase imaging, photonic time stretch, and deep learning enabled by big imaging data analytics. All three components were developed using MATLAB.
The study was led by Bahram Jalali, UCLA professor and Northrop-Grumman Optoelectronics Chair in electrical engineering; Claire Lifan Chen, a UCLA doctoral student; and Ata Mahjoubfar, a UCLA postdoctoral fellow.
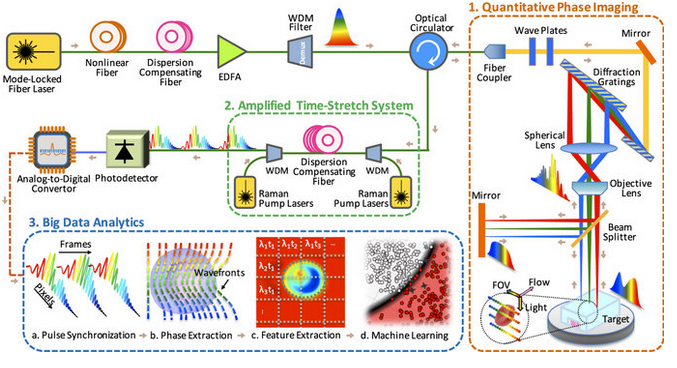
Time-stretch quantitative phase imaging and analytics system. Image credit: Claire Lifan Chen et al./Nature Scientific Reports.
Part 1: Quantitative phase imaging
The microscope uses rainbow pulses of laser light, where distinct colors of light target different locations, resulting in space-to-spectrum encoding. Each pulse lasts less than one billionth of a second. The ultrashort laser pulses freeze the motion of cells, giving blur-free images. The photons with different wavelengths (colors) carry different pieces of information about the cells.
As the photons travel through the cells, the information about the spatial distributions of the cells’ protein concentrations and biomasses are encoded in the phase and amplitude of the laser pulses at various wavelengths. This information is later decoded in the big data analytics pipeline and used by the artificial intelligence algorithm to accurately identify cancerous cells without any need for biomarkers.
The microscope also uses low illumination and post-amplification. This solves the common problem in label-free cell analysis, where the illumination used to capture the images destroys the cells.
“In our system, the average power was a few milliwatts in the field of view, which is very safe for the cells,” stated Claire Lifan Chen. “But, after passing through the cells, we amplify the signal by about 30 times in the photonic time stretch.”
Specially designed optics improve image clarity in this microscope. The researchers have used MATLAB to design the microscope according to the resolution requirements in their biomedical experiments. Image reconstruction and phase recovery are also designed in MATLAB for further downstream analysis.
Part 2: Amplified time-stretch system
Photonic time stretch, invented by Professor Jalali, transforms the optical signals and slows the images so that the microscope can capture them. The microscope can image 100,000 cells per second. The images are digitized at a rate of 36 million images per second, equivalent to 50 billion pixels per second.
The photons travel through a long, specialized fiber and arrive at the sensors during the time between laser pulses. MATLAB was used to determine the parameters for this dispersion compensating fiber.
“The system adds different delays to the pixels within a pulse and feeds them one-by-one, serially, into the photodetector,” said Ata Mahjoubfar.
A software implementation of the physics behind phase stretch transform (PST) as an image feature detection tool from the UCLA Jalali Lab is available for download from MathWorks File Exchange. The code uses Image Processing Toolbox.
The PST code is also available on Github: It has downloaded or viewed more about 27,000 times since May. At the time of this post, it has the third highest star rating of the current MATLAB repositories.
Part 3: Big data analytics using deep learning
Once the microscope captures the images, the team used domain expertise to extract 16 biophysical features of the cells, such as size and density. Recording these features is like documenting the fingerprints of each individual cell. This method yields a high-dimensional dataset which is used to train the machine learning application for identifying cancer cells.
With millions of images to analyze, the team turned to machine learning to solve the multivariate problems. Using MATLAB, the authors implemented and compared the accuracy of multiple machine learning approaches ranging from Naive Bayes and Support Vector Machines, to Deep Neural Network (DNN).
The approach that provided the best performance was a custom DNN implementation that maximized the area under ROC curve (AUC) during training. The AUC is calculated based upon the entire data set, providing inherently high accuracy and repeatability. The success of DNNs for this challenging problem is derived from their ability to learn complex nonlinear relationships between input and output.
In comparing the performance of various classification algorithms, the DNN AUC classifier provided an accuracy of 95.5 % compared to 88.7 % for a simple Naive Bayes classifier. For applications like cancer cell classification, where implications of positive or negative diagnosis are so high, this improvement in accuracy highly impacts the usability of a method.
The more images provided, the more accurate the training system can become. The team trained the machine learning model with millions of line images and used parallel processing to speed up the data analytics. Using high-performance computing (HPC), the image processing and deep learning can be readily performed in real-time by an online learning configuration.
The language of innovation
Not content with simply developing a new microscopy technique, the team utilized the augmented microscope to detect cancer cells more accurately and in less time than current techniques. They also were able to determine which strains of algae provide the most lipids for subsequent refining into biofuels. Yes, one methodology that tackled both biofuels and cancer.
Innovation cannot be achieved without creativity, inspiration, and dedication. Having an interdisciplinary team also helps: This work was a result of collaborations between computer scientists, electrical engineers, bioengineers, chemists, and physicians.
“As we were developing our artificial-intelligence microscope, we crossed the boundaries of scientific fields and modeled biological, optical, electrical, and data analytic systems interconnectedly. MATLAB is a great language for prototyping and provided cohesion to this process, ” said Ata Mahjoubfar.
And, as these researchers demonstrate so eloquently in their publication, having a single language of innovation can facilitate collaboration and accelerate innovation amongst various disciplines in science and engineering.
You can find this and other useful community-developed tools for microscopy here.
For more information on this research, read this article by the UCLA researchers.








Comments
To leave a comment, please click here to sign in to your MathWorks Account or create a new one.