Web Scraping and Mining Unstructured Data with MATLAB
A lot of information is shared on the web and a lot of people are interested in taking advantage of it. It can be used to enrich the existing data, for example. However, information is buries in HTML tags and it is not easy to extract useful information. Today's guest blogger, Toshi Takeuchi shows us how he uses MATLAB for web scraping to harvest useful data from the web and then uses fuzzy string match to enrich existing data.
Contents
- NIPS 2015 Papers
- Scraping Data from a Web Page
- Matching Scraped Data to Database Table
- Fuzzy String Matching
- Validating Fuzzy Match Results
- Updating Missing Values with Fuzzy Match Result
- Reviewing Unmatched
- Substring Match
- Updating Missing Values with Substring Match Result
- Visualizing Paper Author Affiliation
- Summary
NIPS 2015 Papers
Web scraping is actually pretty easy with MATLAB thanks to new string fucntions introdiced in R2016B.
I am going to use as an example the same data used in Text Mining Machine Learning Research Papers with MATLAB.
If you would like to follow along, please download
- the source of this post by clicking on "Get the MATLAB code" at the bottom of this page
- the data from Kaggle's NIPS 2015 Papers page
- my custom function levenshtein.m
- my custom script to generate GIF animation animateNIPS2015.m
Here I am using sqlite in Database Toolbox to load data from a sqlite file. If you don't have Database Toolbox, you can try readtable to read CSV files. Authors table only list names, but I want to enrich it with authors' affilation to see which organizations are active in this academic conference.
db = 'output/database.sqlite'; % database file conn = sqlite(db,'readonly'); % create connection Authors = fetch(conn,'SELECT * FROM Authors'); % get data with SQL command Papers = fetch(conn,'SELECT * FROM Papers'); % get data with SQL command PaperAuthors = fetch(conn,'SELECT * FROM PaperAuthors'); % get data with SQL command close(conn) % close connection Authors = cell2table(Authors,'VariableNames',{'ID','Name'}); % convert to table Papers = cell2table(Papers,'VariableNames', ... % convert to table {'ID','Title','EventType','PdfName','Abstract','PaperText'}); PaperAuthors = cell2table(PaperAuthors,'VariableNames', ... % convert to table {'ID','PaperID','AuthorID'}); head(Authors)
ans =
8×2 table
ID Name
___ ______________________
178 'Yoshua Bengio'
200 'Yann LeCun'
205 'Avrim Blum'
347 'Jonathan D. Cohen'
350 'Samy Bengio'
521 'Alon Orlitsky'
549 'Wulfram Gerstner'
575 'Robert C. Williamson'
Luckily, there is an HTML file that lists each paper with its authors and their affiliation. Each list item start with <i><span class="larger-font"> and ends with </b><br><br>, titles and authors are separated by </span></i><br><b>, mutiple co-authors by semicolon, and finally the names and affiliation by comma.
dbtype output/accepted_papers.html 397:400
397 <div><div><h3>NIPS 2015 Accepted Papers</h3><p><br></p> 398 <i><span class="larger-font">Double or Nothing: Multiplicative Incentive Mechanisms for Crowdsourcing</span></i><br><b> 399 Nihar Shah*, UC Berkeley; Dengyong Zhou, MSR</b><br><br><i><span class="larger-font">Learning with Symmetric Label Noise: The Importance of Being Unhinged</span></i><br><b> 400 Brendan van Rooyen, NICTA; Aditya Menon*, NICTA; Robert Williamson, NICTA</b><br><br><i><span class="larger-font">Algorithmic Stability and Uniform Generalization</span></i><br><b>
Since I have the html file locally, I use fileread to load text from it. If you want to scape a web page directly, you would use webread instead. Imported text is converted into string to take advantage of built-in string functions,
html = string(fileread('output/accepted_papers.html')); % load text from file % html = string(webread('https://nips.cc/Conferences/2015/AcceptedPapers'));
Scraping Data from a Web Page
Usually scraping data from a web page or other unstructured text data sources requires regular expressions and many people find it powerful but very difficult to use. String functions in MATLAB like extractBetween, extractBefore, extractAfter, erase, and replace makes it ridiculously simple!
pattern1 = '<div><h3>NIPS 2015 Accepted Papers</h3><p><br></p>';% start of list pattern2 = '</div> <!--div class="col-xs-12 col-sm-9"-->'; % end of list list = extractBetween(html, pattern1, pattern2); % extract list pattern1 = '<i><span class="larger-font">'; % start of list item pattern2 = '</b><br><br>'; % end of list item listitems = extractBetween(list, pattern1, pattern2); % extract list items pattern1 = ['</span></i><br><b>' newline]; % end of title titles = extractBefore(listitems,pattern1); % extract titles namesorgs = extractAfter(listitems,pattern1); % extract names orgs namesorgs = erase(namesorgs,'*'); % erase * namesorgs = erase(namesorgs,'"'); % erase " namesorgs = replace(namesorgs,' ', ' '); % remove double space disp([titles(1:2) namesorgs(1:2)])
"Double or Nothing: Multiplicati…" "Nihar Shah, UC Berkeley; Dengyo…"
"Learning with Symmetric Label N…" "Brendan van Rooyen, NICTA; Adit…"
Since multiple co-authors are still contained in a single string, let's separate them into a list of co-authors and their affilation. When you split a string, you get varying number of substrings depending on each row. So we need to use arrayfun to parse|UniformOutput| option set to false to split it row by row, trim the result with strtrim the cell aray with vertcat to get the list of coauthors with their affiliation.
namesorgs = replace(namesorgs,'&','&'); % revert escaped & namesorgs = erase(namesorgs,[char(194) ' </b><span><strong>']); % remove extra tags namesorgs = replace(namesorgs,['</strong></span><b>' ... % replace missing semicolon char(194)],';'); coauth = arrayfun(@(x) strtrim(split(x,';')), namesorgs, ... % split by semicolon 'UniformOutput', false); % and trim white space coauth = vertcat(coauth{:}); % unnest cell array coauth(1:5)
ans =
5×1 string array
"Nihar Shah, UC Berkeley"
"Dengyong Zhou, MSR"
"Brendan van Rooyen, NICTA"
"Aditya Menon, NICTA"
"Robert Williamson, NICTA"
Matching Scraped Data to Database Table
You now see how easy web scraping is with MATLAB.
Now that we have the list of names with affiliation, we just have to match it to the Authors table by name, right? Unfortunately, we see a lot of missing values because the names didn't match even with contains partial match function.
The real hard part now is what to do after you scraped the data from the web.
authors = Authors.Name; % author names names = strtrim(extractBefore(coauth,',')); % extract and trim names org = strings(length(authors),1); % initialize accumulator for ii = 1:length(authors) % for each name in |authors| res = coauth(contains(names,authors(ii))); % find match in |names| if isempty(res) % if no match org(ii) = missing; % mark it missing end res = extractAfter(res,','); % extract after comma res = strtrim(res); % remove white space res = unique(res); % remove duplicates res(strlength(res) == 0) = []; % remove emptry string if length(res) == 1 % if single string org(ii) = res; % use it as is elseif length(res) > 1 % if multiple string org(ii) = join(res,';'); % join them with semicolon else % otherwise org(ii) = missing; % mark it missing end end head(table(authors, org, 'VariableNames',{'Name','Org'}))
ans =
8×2 table
Name Org
______________________ _____________________________________
'Yoshua Bengio' "U. Montreal"
'Yann LeCun' "New York University"
'Avrim Blum' <missing>
'Jonathan D. Cohen' <missing>
'Samy Bengio' "Google Research"
'Alon Orlitsky' "University of California, San Diego"
'Wulfram Gerstner' "EPFL"
'Robert C. Williamson' <missing>
For example, the partial match doesn't work if middle initial is missing or nicknames are used instead of full names. There can be other irregularities. Yikes, we are dealing with unstructured data!
[authors(4) coauth(contains(names,'Jonathan Cohen')); authors(8) coauth(contains(names,'Robert Williamson')); authors(406) coauth(contains(names,'Sanmi Koyejo')); authors(440) coauth(contains(names,'Danilo Rezende')); authors(743) coauth(contains(names,'Bill Dally')); authors(769) coauth(contains(names,'Julian Yarkony'))]
ans =
6×2 string array
"Jonathan D. Cohen" "Jonathan Cohen, Princeton Unive…"
"Robert C. Williamson" "Robert Williamson, NICTA"
"Oluwasanmi O. Koyejo" "Sanmi Koyejo, Stanford University"
"Danilo Jimenez Rezende" "Danilo Rezende, Google DeepMind"
"William Dally" "Bill Dally , Stanford Universit…"
"Julian E. Yarkony" "Julian Yarkony, Dr."
Fuzzy String Matching
What can we do when exact match approach doesn't work? Maybe we can come up with various rules to match strings using regular expressions, but that is very time consuming. Let's revisit the 40-year-old Algorithm That Cannot Be Improved to solve this problem. I created a new custom function levenshtein for this example. It calculates the edit distance that measures the minimum number of edit operations required to transform one string into another,as a way to quantify how similar or dissimilar they are. For more details of this algorithm, please check out the blog post linked above
Converting Sunday to Saturday requires 3 edit operations.
levenshtein('sunday', 'saturday')
ans =
3
Perhaps it is easier to understand if I show how similar they are as match rate rather than number of edit operations?
levenshtein('sunday', 'saturday', 'ratio')
ans =
0.78571
Now we can find "Jonathan Cohen" in the top 3 matches for "Jonathan D. Cohen".
fhandle = @(s) levenshtein(authors(4), s, 'ratio'); % function handle ratios = arrayfun(fhandle, extractBefore(coauth,',')); % get match rates [~,idx] = sort(ratios,'descend'); % rank by match rate [repmat(authors(4),[3,1]) coauth(idx(1:3)) ratios(idx(1:3))]
ans =
3×3 string array
"Jonathan D. Cohen" "Jonathan Cohen, Pr…" "0.90323"
"Jonathan D. Cohen" "Jonathan Bassen, s…" "0.8125"
"Jonathan D. Cohen" "Jonathan Vacher, U…" "0.8125"
Validating Fuzzy Match Results
Let;s try this approach for the first 10 missing names with ignoreCase option enabled. It looks like we can be fairly confident about the result as long as the maximum match rate is 0.8 or higher.
org(org == 'Dr.') = missing; % remove salutation missing_ids = find(ismissing(org)); % get missing ids matches = strings(1,3); % initialize accumulator for ii = 1:10 % for each missing id cid = missing_ids(ii); % current id fhandle = @(s) levenshtein(authors(cid), s, 'ratio', 'ingoreCase'); ratios = arrayfun(fhandle, names); % get match rates [~,idx] = max(ratios); % get index of max value matches(ii,:) = [authors(cid) coauth(idx) ratios(idx)]; % update accumulator end matches
matches =
10×3 string array
"Avrim Blum" "Manuel Blum, Unive…" "0.71429"
"Jonathan D. Cohen" "Jonathan Cohen, Pr…" "0.90323"
"Robert C. Williamson" "Robert Williamson,…" "0.91892"
"Juergen Schmidhuber" "J?rgen Schmidhuber," "0.94595"
"Wolfgang Maass" "Wolfgang Maass," "1"
"Robert E. Schapire" "Robert Schapire, M…" "0.90909"
"Tomás Lozano-Pérez" "Tomás Lozano-Pér…" "1"
"Dit-Yan Yeung" "Dit Yan Yeung, HKUST" "0.96154"
"Geoffrey J. Gordon" "Geoff Gordon, CMU" "0.8"
"Brendan J. Frey" "Brendan Frey, U. T…" "0.88889"
Updating Missing Values with Fuzzy Match Result
Now we can apply this approach to update the missing values. Now 89.3% of missing values are identified!
for ii = 1:length(missing_ids) % for each missing id cid = missing_ids(ii); % current id fhandle = @(s) levenshtein(authors(cid), s, 'ratio', 'ignoreCase'); ratios = arrayfun(fhandle, names); % get match rates [~,idx] = max(ratios); % get index of max value if ratios(idx) >= 0.8 % if max is 0.8 res = extractAfter(coauth(idx),','); % get org name res = strtrim(res); % trim white spaces if strlength(res) == 0 % if null string org(cid) = 'UNK'; % unknown else % otherwise org(cid) = res; % update org end end end fprintf('Missing reduced by %.1f%%\n', (1 - sum(ismissing(org))/length(missing_ids))*100)
Missing reduced by 89.3%
Reviewing Unmatched
When you review what was not matched, you can see that they were no good matches except for "John W. Fisher III", "Oluwasanmi O. Koyejo", or "Danielo Jimenz Rezende". They have much lower match rate and didn't make the cut, but we don't necessarily ease the cut-off. Is there an alternative?
missing_ids = find(ismissing(org)); % get missing ids matches = strings(1,3); % initialize accumulator for ii = 1:10 % for each missing id thru 10th cid = missing_ids(ii); % current id fhandle = @(s) levenshtein(authors(cid), s, 'ratio', 'ignoreCase'); ratios = arrayfun(fhandle, names); % get match rates [~,idx] = max(ratios); % get index of max value matches(ii,:) = [authors(cid) coauth(idx) ratios(idx)]; % update accumulator end matches
matches =
10×3 string array
"Avrim Blum" "Manuel Blum, Unive…" "0.7619"
"John W. Fisher III" "John Fisher, MIT" "0.75862"
"Özgür ÅžimÅŸek" "Ozgur Simsek, Max …" "0.71429"
"Joseph J. Lim" "Joseph Salmon, Tel…" "0.76923"
"Pascal Fua" "Pascal Vincent, U.…" "0.70833"
"François Fleuret" "Matthias Feurer, U…" "0.71875"
"Oluwasanmi O. Koyejo" "Sanmi Koyejo, Stan…" "0.75"
"Samet Oymak" "James Kwok, Hong K…" "0.71429"
"Danilo Jimenez Rez…" "Danilo Rezende, Go…" "0.77778"
"Kafui Dzirasa" "Rahul Krishnan, Ne…" "0.66667"
Substring Match
Instead of trying to match the whole string, we can reorder and find the most similar substring with the longer string based on the shorter string using ignore order and partial options:
- ignoreOrder breaks strings into tokens, reorder tokens by finding union and intersection of tokens, and compute edit distance.
- partial finds substring within longer string that is closest to the shorter string.
This clearly gives higher match rates for in some specific cases. "John W. Fisher III" can be reordered to "John Fisher III W." and the first two tokens (a substring) matches "John Fisher", which makes it a 100% match.
matches = strings(1,3); % initialize accumulator for ii = 1:10 % for each missing id thru 10th cid = missing_ids(ii); % current id fhandle = @(s) levenshtein(authors(cid), s, 'ratio', 'ingoreCase', 'ignoreOrder', 'partial'); ratios = arrayfun(fhandle, names); % get match rates [~,idx] = max(ratios); % get index of max value matches(ii,:) = [authors(cid) coauth(idx) ratios(idx)]; % update accumulator end matches
matches =
10×3 string array
"Avrim Blum" "Manuel Blum, Unive…" "0.75"
"John W. Fisher III" "John Fisher, MIT" "1"
"Özgür ÅžimÅŸek" "Ozgur Simsek, Max …" "0.66667"
"Joseph J. Lim" "Joseph Wang," "0.81818"
"Pascal Fua" "Pascal Vincent, U.…" "0.85"
"François Fleuret" "Tian Tian, Tsinghu…" "0.75"
"Oluwasanmi O. Koyejo" "Sanmi Koyejo, Stan…" "0.79167"
"Samet Oymak" "James Hensman, The…" "0.77273"
"Danilo Jimenez Rez…" "Danilo Rezende, Go…" "1"
"Kafui Dzirasa" "Kai Fan, Duke Univ…" "0.71429"
Updating Missing Values with Substring Match Result
Substring match is a tricky choice - it can produce more false positives.So let's use match rate of 0.9 or above for cut-off. With this approach, the missing values were further reduced by 35.5%.
for ii = 1:length(missing_ids) % for each missing id cid = missing_ids(ii); % current id fhandle = @(s) levenshtein(authors(cid), s, 'ratio', 'ingoreCase', 'ignoreOrder', 'partial'); ratios = arrayfun(fhandle, names); % get match rates [~,idx] = max(ratios); % get index of max value if ratios(idx) >= 0.9 % if max is 0.9 res = extractAfter(coauth(idx),','); % get org name res = strtrim(res); % trim white spaces if strlength(res) == 0 % if null string org(cid) = 'UNK'; % unknown else % otherwise org(cid) = res; % update org end end end fprintf('Missing reduced by %.1f%%\n', (1 - sum(ismissing(org))/length(missing_ids))*100)
Missing reduced by 35.5%
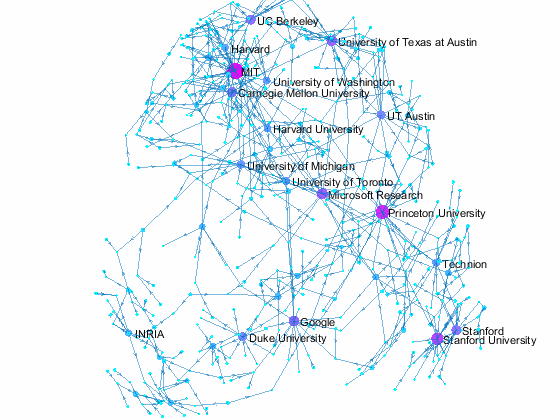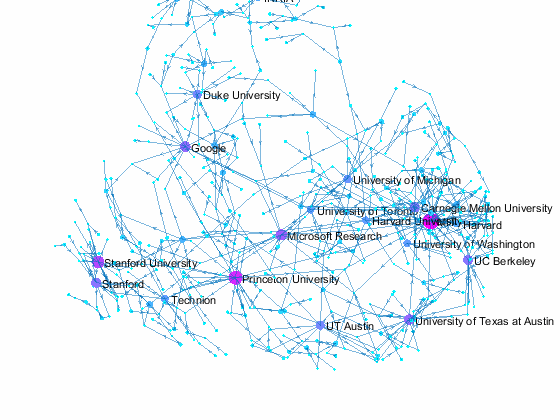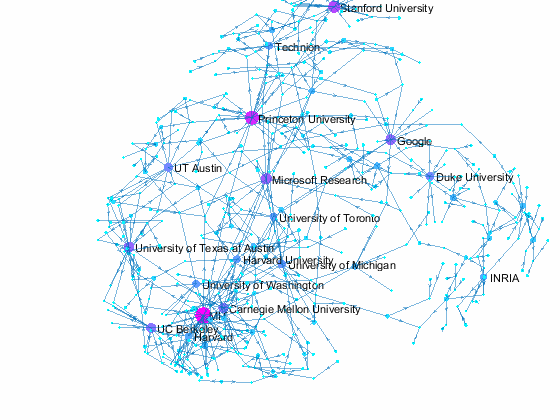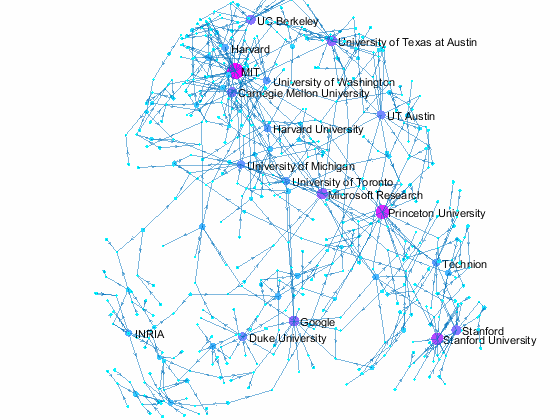
Visualizing Paper Author Affiliation
Now we can use the matched data to visualize the graph formed by Paper Author Affiliation. PaperAuthor table can be seen as the edge list where paper and authors represents nodes of a graph and each row represents an edge. We just need to replace authors with their affiliation so that each node represents a paper or the affiliation of its authors, and edges represents the authorship of the paper.
- A paper has multiple incoming edges if co-authored by scholars from multiple organizations,
- and those papers are also connected to other node if their co-authors also contribute to other papers.
- Papers authored within the same organization will be isolated, and it is removed from the graph for simplifying visualzation.
- The resulting graph represents the largest connected component of the overall graph.
- Node size and color reflects outdegrees - how many papers is contributed by the members of a given organization
This visualization shows that we have more work to do - for example, "University of Texas Austin" and "UT Austin" are the same organization but we have two separate nodes because organizatoin names are not standardized, and that means some organizations are probably under-reprsented. Can you improve this visualzation?
Af = table(Authors.ID,org, 'VariableNames',{'AuthorID','Org'}); % affiliation Af(Af.Org == 'UNK' | ismissing(Af.Org),:) = []; % remove UNK & missing T = innerjoin(PaperAuthors(:,2:3), Af); % join with PaperAuthors T = innerjoin(T,Papers(:,[1,2]), 'Keys', 1); % join with Papers [T,~,idx] = unique(T(:,[3,4]),'rows'); % remove duplicate rows w = accumarray(idx,1); % count duplicates s = cellstr(T.Org); % convert to cellstr t = cellstr(T.Title); % convert to cellstr G = digraph(s,t, w); % create directed graph bins = conncomp(G,'type', 'weak', 'OutputForm', 'cell'); % get connected comps [~, idx] = max(cellfun(@length, bins)); % find largest comp G = subgraph(G, bins{idx}); % subgraph largest comp figure % new figure colormap cool % use cool colormap msize = 10*(outdegree(G) + 3)./max(outdegree(G)); % marker size ncol = outdegree(G) + 3; % node colors named = outdegree(G) > 7; % nodes to label h = plot(G, 'MarkerSize', msize, 'NodeCData', ncol); % plot graph layout(h,'force3','Iterations',30) % change layout labelnode(h, find(named), G.Nodes.Name(named)); % add node labels title('NIPS 2015 Papers - Author Affilation Graph') % add title axis tight off % set axis set(gca,'clipping','off') % turn off clipping zoom(1.3) % zoom in

Summary
Web scraping can be a very useful skill to have to collect information from the web, and MATLAB makes it very easy to extract information from a web page. The resulting data is often unstructured, but you can deal with it using techniques like fuzzy string matching.
I hope this example gives you a lot of new ideas. Give it a try and let us know how your experience goes here!








评论
要发表评论,请点击 此处 登录到您的 MathWorks 帐户或创建一个新帐户。