Using Ensemble Learning to Create Accurate Machine Learning Algorithms
In today’s post, Grace from the Student Programs Team will show how you can started with ensemble learning. Over to you, Grace!
When building a predictive machine learning model, there are many ways to improve it’s performance: try out different algorithms, optimize the parameters of the algorithm, find the best way to divide and process your data, and more. Another great way to create accurate predictive models is through ensemble learning.
What is ensemble learning?
Ensemble learning is the practice of combining multiple machine learning models into one predictive model. Some types of machine learning algorithms are considered weak learners, meaning that they are highly sensitive to the data that is used to train them and are prone to inaccuracies. Creating an ensemble of weak learners and aggregating their results to make predictions on new observations often results in a single higher-quality model. At it’s simplest, ensemble learning can be represented with the animation below:
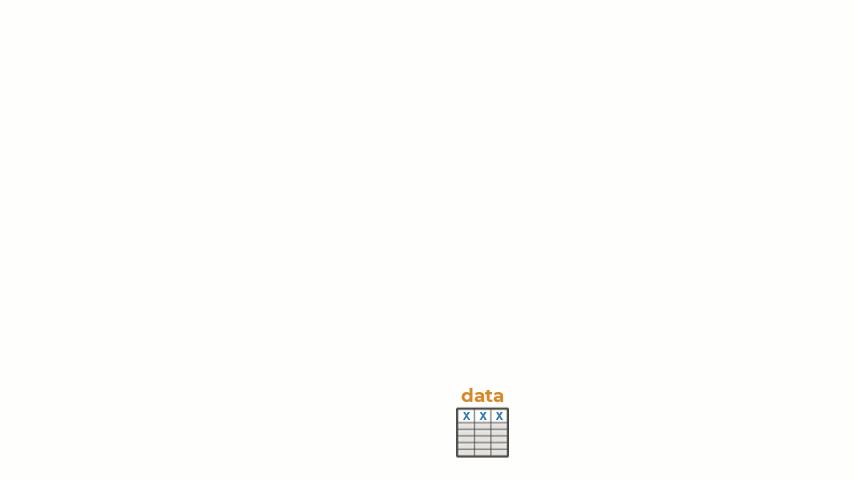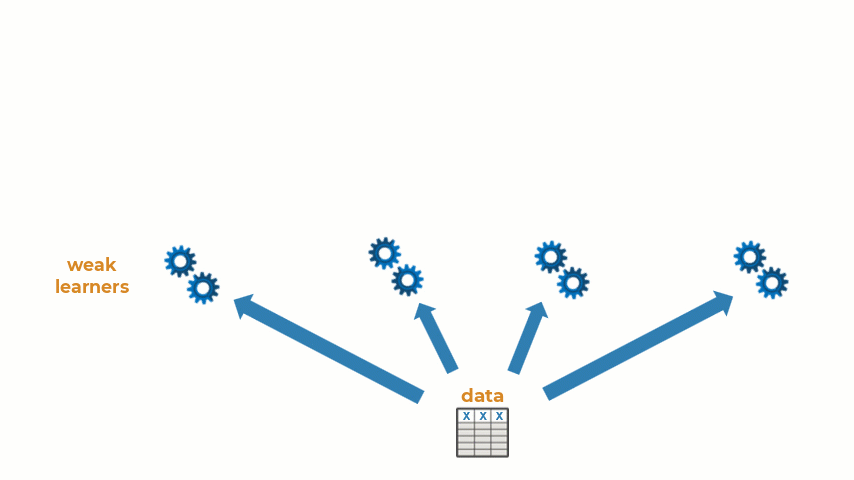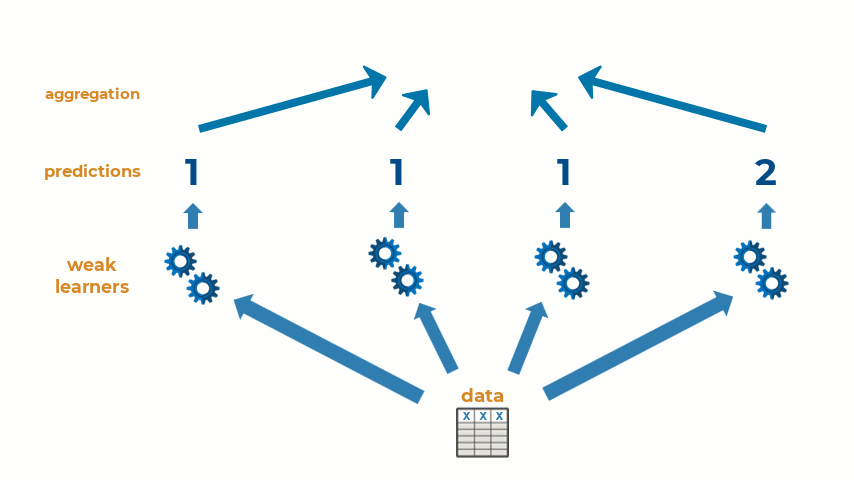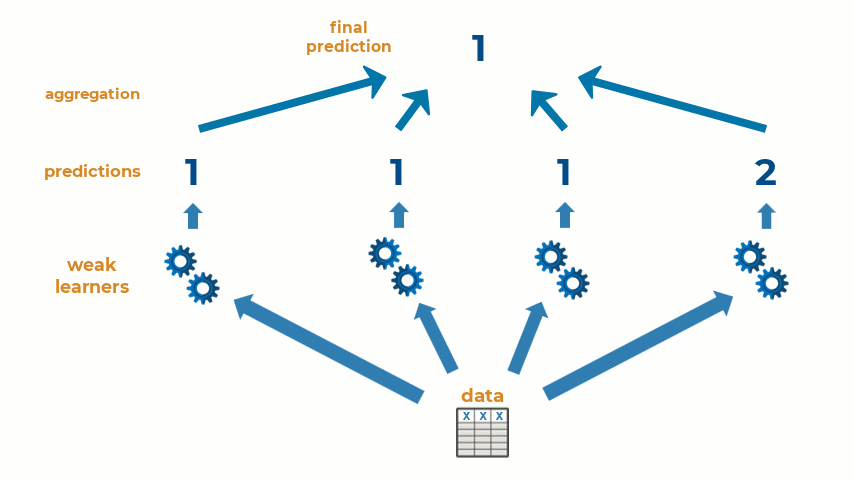
Ensemble learning can be used for a wide variety of machine and deep learning methods. Today, I will show how to create an ensemble of machine learning models for a regression problem, though the workflow will be similar for classification problems as well. Let’s get started!
1. Prepare the data
For this problem, we have a set of tabular data pertaining to cars, as shown below:
load carbig
carTable = table(Acceleration,Cylinders,Displacement, …
Horsepower,Model_Year,Weight,MPG);
head(carTable)
Our goal is to create a model that can accurately predict what a car’s mileage per gallon (MPG) will be. With any data problem, you should take the time to explore and preprocess the data, but for this tutorial I will just do some simple steps. For more information on cleaning your data, check out this example that shows a lot of great ways you can preprocess tabular data!
First, I’ll check if our set has any rows with missing data, as this will inform some of our decisions later.
missingElements = ismissing(carTable);
rowsWithMissingValues = any(missingElements,2);
missingValuesTable = carTable(rowsWithMissingValues,:)
There are a total of 14 rows with missing data, 8 of which are missing the ‘MPG’ value. I’ll remove these rows, as they are not helpful for training, but we will use the other rows as they could still provide helpful information when training.
rowsMissingMPG = ismissing(carTable.MPG);
carTable(rowsMissingMPG,: ) = []
Last, I will split our data into a training set and a testing set, which will be used to teach and evaluate the ensemble, respectively. Using the dividerand function, I put 70% of the data into the training set and 30% into the testing set as a starting point, but you can test out different divisions of data when building your own models.
numRows = size(carTable,1);
[trainInd, ~, testInd] = dividerand(numRows, .7, 0, .3);
trainingData = carTable(trainInd, :);
testingData = carTable(testInd, :);
2. Create an ensemble
Now that our data is ready, it’s time to start creating the ensemble! I’ll start by showing the easiest way to create an ensemble using the default parameters for each individual learner, and then I’ll also show how to use templates to customize your weak learners.
Using Bulit-In Algorithms
You can create an ensemble for regression by using fitrensemble (fitcensemble for classification). With just this function and your data, you could have an ensemble ready by executing the following line of code:
Mdl = fitrensemble(trainingData, ‘MPG’);
This will use all the default settings for training an ensemble of weak regression learners: 100 trees are trained and they are aggregated using the least-squares boosting (LSBoost) algorithm.
However, fitrensemble also provides the option to customize the ensemble settings, so I will specify a few of these settings. First, I want to use the ‘Bag’ method of aggregation instead of the ‘LSBoost’ method because it tends to have higher accuracy and our dataset is relatively small. For a full list of aggregation algorithms and some suggestions on how to choose a starting algorithm, check out this documentation page!
I also want to specify how many learners will be in the ensemble, which is set by the ‘NumLearningCycles’ property. To choose how many learners the ensemble will have, try starting with several dozen, training the ensemble, and then checking the ensemble quality. If the ensemble is not accurate and could benefit from more learners, then you can adjust this number or add them in later! For now, I’ll start with 30 learners.
Both of these options are set using Name-Value arguments, as shown below.
Mdl = fitrensemble(trainingData, ‘MPG’, ‘Method’, ‘Bag’, ‘NumLearningCycles’, 30);
And just like that, we’ve trained an ensemble of 50 learners that are ready to be used on new data!
Using Templates
There may be times when you want to change some parameters of the individual learners, not just of the ensemble. To do that, we can use learner templates.
Unless otherwise specified, fitrensemble creates an ensemble of default regression tree learners, but this may not always be what you want. As we saw earlier, our data has some missing values, which can decrease the performance of these trees. Trees that use surrogate splits tend to perform better with missing data than trees that don’t, so I will use the templateTree function to specify that I want the learners to use surrogate splits.
templ = templateTree(‘Surrogate’,‘all’, ‘Type’, ‘regression’);
templMdl = fitrensemble(trainingData, ‘MPG’, ‘Method’, ‘Bag’, ‘NumLearningCycles’, 30, ‘Learners’, templ);
As before, we end up with an ensemble that can be used to make predictions on new data! While you can give fitcensemble and fitrensemble a cell array of learner templates, the most common usage is to give just one weak learner template.
3. Evaluate the Ensemble
Once you have a trained ensemble, it’s time to see how well it performs! The predictive quality of an ensemble cannot be evaluated based on its performance on training data, as the ensemble knows it too well. It’s very likely that it will perform really well on the training data, but that does not mean it will perform well on any other data. To obtain a better idea of the quality of an ensemble, you can use one of these methods:
- If you DON’T have a lot of training data: Evaluate the ensemble by cross validation
- If you DO have a lot of training data: Evaluate the ensemble on an independent test set
I will show how to use both of these methods to evaluate a model in the following sections.
Evaluate through Cross-Validation
Cross validation is a common technique for evaluating a model’s performance by partitioning the total dataset and using some partitions for training and others for testing. There are multiple types of cross-validation, and many allow you to eventually use all of the training data to train the model, which is what makes it ideal for smaller datasets. If you are not familiar with cross-validation, check out this discovery page to learn more!
For this example, I will be using k-fold cross-validation. You can perform cross-validation when creating your ensemble by using the ‘CrossVal’ Name-Value Argument of fitrensemble, as outlined below:
Mdl = fitrensemble(trainingData, ‘MPG’, ‘CrossVal’, ‘on’);
Since we already have an ensemble, however, we can use the crossval function to cross-validate our model, then use kfoldLoss to extract the average mean squared error (MSE), or loss, of our final model.
cvens = crossval(templMdl);
kfoldLoss(cvens)
We can also set the ‘mode’ of kfoldLoss to ‘cumulative’ and then plot the results to show how the loss value changes as more trees are trained.
cValLoss = kfoldLoss(cvens,‘mode’,‘cumulative’)
plot(cValLoss, ‘r–‘)
xlabel(‘Number of trees’)
ylabel(‘Cross-Validation loss’)
Evaluate on test set
If you have enough data to use only a portion of it for training, you can use the rest of the data to test how well your model performs. First, make sure you separate your data into a training and testing set, as we did earlier, then train your model using only the training set.
Once the ensemble is trained, we can use it on the testing data and then calculate the loss of the model on this data:
loss(templMdl,testingData,“MPG”)
plot(loss(templMdl,testingData,“MPG”,‘mode’,‘cumulative’))
xlabel(‘Number of trees’)
ylabel(‘Test loss’)
You can also use the ensemble to make predictions using the predict function. With a test set, you can compare the expected results from the testing data to the results predicted by the ensemble. In the plot below, the blue line represents the expected results, and the red circles are the predicted results; the further away a circle is from the blue line, the less accurate the prediction was.
predMPG = predict(templMdl, testingData);
expectMPG = testingData.MPG;
plot(expectMPG, expectMPG);
hold on
scatter(expectMPG, predMPG)
xlabel(‘True Response’)
ylabel(‘Predicted Response’)
hold off
You can use these evaluation metrics to compare multiple ensembles and choose the one that performs the best.
4. Iterate and Improve!
As with any machine learning workflow, it’s important to try out different algorithms until you get an ensemble that you are happy with. When I first started creating this ensemble, I used the ‘LSBoost’ aggregation method instead of ‘Bag’ and the performance was consistently pretty poor, so I changed this property in line 17 (and 19) and re-ran the entire Live Script, resulting in a new, fully evaluated model in a matter of seconds. In addition to testing out different aggregation algorithms, here are some other suggestions for improving your ensemble:
- If it appears that the loss of your ensemble is still decreasing when all members have finished training, this could indicate that you need more members. You can add them using the resume method. Repeat until adding more members does not improve ensemble quality.
- Try optimizing your hyperparameters automatically by using the ‘OptimizeHyperparameters’ and ‘HyperparameterOptimizationOptions’ Name-Value arguments when calling fitrensemble. Check out this example in the documentation to learn more: Hyperparameter optimization.
- Test out different weak learners! There are lots of different settings and templates you can use, especially if you’re creating a classification ensemble. Try different parameters when calling fitrensemble or fitcensemble, use different template types, and play around with the different options of each template.
- At the end of the day, a model is only as good as the data it is trained on, so make sure your data is clean and test out different divisions of training and testing data to see what works best for your ensemble. There are many different ways to clean data depending on what format it is in, so use this documentation page as a starting point to find resources based on the format and patterns of your dataset!
If you are interested in deep learning and would like to learn about ensemble learning with neural networks, check out this blog post next!
- Category:
- Data Science












Comments
To leave a comment, please click here to sign in to your MathWorks Account or create a new one.