Subset Selection and Regularization
This week Richard Willey from technical marketing will be guest blogging about subset selection and regularization. This week's blog posting is motivated by a pair of common challenges that occur in applied curve fitting.
- The first challenge is how best to create accurate predictive models when your independent variables exhibit strong correlation. In many cases you can improve upon the results of an ordinary least square regression if you reduce the number of predictors or, alternatively, shrink the coefficient values towards zero.
- The second challenge involves generating an accurate predictive model when you are constrained with respect to the number of predictors you are working with. For example, assume that you need to embed your model onto a controller. You have 20 possible predictors to chose from, but you only have enough memory to allow for 8 independent variables.
This blog post will show two different sets of techniques to solve these related challenges. The first set of techniques are based on a combination of feature selection and cross validation. The second set of techniques are use regularization algorithms like ridge regression, lasso and the elastic net. All of these algorithms can be found in Statistics Toolbox.
Contents
Create a Dataset
To start with, I am going to generate a dataset that is deliberately engineered to be very difficult to fit using traditional linear regression. The dataset has three characteristics that present challenges of linear regression
- The dataset is very "wide". There are relatively large numbers of (possible) predictors compared to the number of observations.
- The predictors are strongly correlated with one another
- There is a large amount of noise in the dataset
clear all
clc
rng(1998);Create eight X variables. The mean of each variable will be equal to zero.
mu = [0 0 0 0 0 0 0 0];
The variables are correlated with one another. The covariance matrix is specified as
i = 1:8; matrix = abs(bsxfun(@minus,i',i)); covariance = repmat(.5,8,8).^matrix;
Use these parameters to generate a set of multivariate normal random numbers.
X = mvnrnd(mu, covariance, 20);
Create a hyperplane that describes Y = f(X) and add in a noise vector. Note that only three of the eight predictors have coefficient values > 0. This means that five of the eight predictors have zero impact on the actual model.
Beta = [3; 1.5; 0; 0; 2; 0; 0; 0]; ds = dataset(Beta); Y = X * Beta + 3 * randn(20,1);
For those who are interested, this example was motivated by Robert Tibshirani's classic 1996 paper on the lasso:
Tibshirani, R. (1996). Regression shrinkage and selection via the lasso. J. Royal. Statist. Soc B., Vol. 58, No. 1, pages 267-288).
Let's model Y = f(X) using ordinary least squares regression. We'll start by using traditional linear regression to model Y = f(X) and compare the coefficients that were estimated by the regression model to the known vector Beta.
b = regress(Y,X); ds.Linear = b; disp(ds)
Beta Linear
3 3.5819
1.5 0.44611
0 0.92272
0 -0.84134
2 4.7091
0 -2.5195
0 0.17123
0 -0.42067
As expected, the regression model is performing very poorly.
- The regression is estimating coefficient values for all eight predictors. Objectively, we "know" that five of these coefficients should be zero.
- Furthermore, the coefficients that are being estimated are very far removed from the true value.
Let's see if we can improve on these results using a technique called "All possible subset regression". First, I'm going to generate 255 different regession models which encompass every possible combinate of the eight predictors. Next, I am going to measure the accuracy of the resulting regression using the cross validated mean squared error. Finally, I'm going to select the regression model that produced the most accurate results and compare this to our original regression model.
Create an index for the regression subsets.
index = dec2bin(1:255);
index = index == '1';
results = double(index);
results(:,9) = zeros(length(results),1);Generate the regression models.
for i = 1:length(index) foo = index(i,:); rng(1981); regf = @(XTRAIN, YTRAIN, XTEST)(XTEST*regress(YTRAIN,XTRAIN)); results(i,9) = crossval('mse', X(:,foo), Y,'kfold', 5, 'predfun',regf); end
Sort the outputs by MSE. I'll show you the first few.
index = sortrows(results, 9); index(1:10,:)
ans =
Columns 1 through 5
1 1 0 0 1
1 1 0 1 1
1 0 1 0 1
1 1 0 0 1
1 0 0 0 1
1 1 0 1 1
1 0 0 1 1
1 0 0 0 1
1 0 1 0 1
1 0 0 1 1
Columns 6 through 9
1 0 0 10.239
1 0 0 10.533
1 0 0 10.615
0 0 0 10.976
1 0 0 11.062
0 0 0 11.257
1 0 0 11.641
0 0 0 11.938
0 0 0 12.068
0 0 0 12.249
Compare the Results
beta = regress(Y, X(:,logical(index(1,1:8)))); Subset = zeros(8,1); ds.Subset = Subset; ds.Subset(logical(index(1,1:8))) = beta; disp(ds)
Beta Linear Subset
3 3.5819 3.4274
1.5 0.44611 1.1328
0 0.92272 0
0 -0.84134 0
2 4.7091 4.1048
0 -2.5195 -2.0063
0 0.17123 0
0 -0.42067 0
The results are rather striking.
- Our new model only includes 4 predictors. Variables 3,4, 7 and 8 have been eliminated from the regression model. Furthermore, the variables that were exluded are ones that we "know" should be zeroed out.
- Equally significant, the coefficients that are being estimated from the resulting regression model are much closer to the "true" values.
All possible subset regression appears to have generated a significantly better model. This R^2 value for this regression model isn't as good as the original linear regression; however, if we're trying to generate predictions from a new data set, we'd expect this model to perform significantly better.
Perform a Simulation
It's always dangerous to rely on the results of a single observation. If we were to change either of the noise vectors that we used to generate our original dataset we might end up with radically different results. This next block of code will repeat our original experiment a hundred times, each time adding in different noise vectors. Our output will be a bar chart that shows how frequently each of the different variables was included in the resulting model. I'm using parallel computing to speed things up though you could perform this (more slowly) in serial.
matlabpool open sumin = zeros(1,8); parfor j=1:100 mu = [0 0 0 0 0 0 0 0]; Beta = [3; 1.5; 0; 0; 2; 0; 0; 0]; i = 1:8; matrix = abs(bsxfun(@minus,i',i)); covariance = repmat(.5,8,8).^matrix; X = mvnrnd(mu, covariance, 20); Y = X * Beta + 3 * randn(size(X,1),1); index = dec2bin(1:255); index = index == '1'; results = double(index); results(:,9) = zeros(length(results),1); for k = 1:length(index) foo = index(k,:); regf = @(XTRAIN, YTRAIN, XTEST)(XTEST*regress(YTRAIN,XTRAIN)); results(k,9) = crossval('mse', X(:,foo), Y,'kfold', 5, ... 'predfun',regf); end % Sort the outputs by MSE index = sortrows(results, 9); sumin = sumin + index(1,1:8); end % parfor matlabpool close
Starting matlabpool using the 'local4' configuration ... connected to 3 labs. Sending a stop signal to all the labs ... stopped.
%Plot results.
bar(sumin)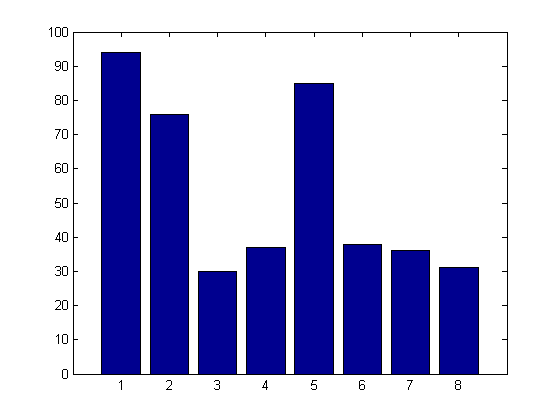
The results clearly show that "All possible subset selection" is doing a really good job identifying the key variables that drive our model. Variables 1, 2, and 5 show up in most of the models. Individual distractors work their way into the models as well, however, their frequency is no where near as high as the real predictors.
Introducing Sequential Feature Selection
All possible subset selection suffers from one enormous limitation. It's computationally infeasible to apply this technique to datasets that contain large numbers of (potential) predictors. Our first example had eight possible predictors. Testing all possible subsets required generate (2^8)-1 = 255 different models. Suppose that we were evaluating a model with 20 possible predictors. All possible subsets would require use to test 1,048,575 different models. With 40 predictors we'd be looking at a billion different combinations.
Sequential feature selection is a more modern approach that tries to define a smart path through the search space. In turn, this should allow us to identify a good set of cofficients but ensure that the problem is still computationally feasible.
Sequential feature selection works as follows:
- Start by testing each possible predictor one at a time. Identify the single predictor that generates the most accurate model. This predictor is automatically added to the model.
- Next, one at a time, add each of the remaining predictors to a model that includes the single best variable. Identify the variable that improves the accuracy of the model the most.
- Test the two models for statistical significance. If the new model is not significantly more accurate that the original model, stop the process. If, however, the new model is statistically more significant, go and search for the third best variable.
- Repeat this process until you can't identify a new variable that has a statistically significant impact on the model.
% This next bit of code will show you how to use sequential feature % selection to solve a significantly larger problem.
Generate a Data Set
clear all
clc
rng(1981);
Z12 = randn(100,40);
Z1 = randn(100,1);
X = Z12 + repmat(Z1, 1,40);
B0 = zeros(10,1);
B1 = ones(10,1);
Beta = 2 * vertcat(B0,B1,B0,B1);
ds = dataset(Beta);
Y = X * Beta + 15*randn(100,1);Use linear regression to fit the model.
b = regress(Y,X); ds.Linear = b;
Use sequential feature selection to fit the model.
opts = statset('display','iter'); fun = @(x0,y0,x1,y1) norm(y1-x1*(x0\y0))^2; % residual sum of squares [in,history] = sequentialfs(fun,X,Y,'cv',5, 'options',opts)
Start forward sequential feature selection:
Initial columns included: none
Columns that can not be included: none
Step 1, added column 26, criterion value 868.909
Step 2, added column 31, criterion value 566.166
Step 3, added column 9, criterion value 425.096
Step 4, added column 35, criterion value 369.597
Step 5, added column 16, criterion value 314.445
Step 6, added column 5, criterion value 283.324
Step 7, added column 18, criterion value 269.421
Step 8, added column 15, criterion value 253.78
Step 9, added column 40, criterion value 242.04
Step 10, added column 27, criterion value 234.184
Step 11, added column 12, criterion value 223.271
Step 12, added column 17, criterion value 216.464
Step 13, added column 14, criterion value 212.635
Step 14, added column 22, criterion value 207.885
Step 15, added column 7, criterion value 205.253
Step 16, added column 33, criterion value 204.179
Final columns included: 5 7 9 12 14 15 16 17 18 22 26 27 31 33 35 40
in =
Columns 1 through 12
0 0 0 0 1 0 1 0 1 0 0 1
Columns 13 through 24
0 1 1 1 1 1 0 0 0 1 0 0
Columns 25 through 36
0 1 1 0 0 0 1 0 1 0 1 0
Columns 37 through 40
0 0 0 1
history =
In: [16x40 logical]
Crit: [1x16 double]
I've configured sequentialfs to show each iteration of the feature selection process. You can see each of the variables as it gets added to the model, along with the change in the cross validated mean squared error.
Generate a linear regression using the optimal set of features
beta = regress(Y,X(:,in)); ds.Sequential = zeros(length(ds),1); ds.Sequential(in) = beta
ds =
Beta Linear Sequential
0 -0.74167 0
0 -0.66373 0
0 1.2025 0
0 -1.7087 0
0 1.6493 2.5441
0 1.3721 0
0 -1.6411 -1.1617
0 -3.5482 0
0 4.1643 2.9544
0 -1.9067 0
2 0.28763 0
2 1.7454 2.9983
2 0.1005 0
2 2.2226 3.5806
2 4.4912 4.0527
2 4.7129 5.3879
2 0.9606 1.9982
2 5.9834 5.1789
2 3.9699 0
2 0.94454 0
0 2.861 0
0 -2.1236 -2.4748
0 -1.6324 0
0 1.8677 0
0 -0.51398 0
0 3.6856 4.0398
0 -3.3103 -4.1396
0 -0.98227 0
0 1.4756 0
0 0.026193 0
2 2.9373 3.8792
2 0.63056 0
2 2.3509 1.9472
2 0.8035 0
2 2.9242 4.35
2 0.4794 0
2 -0.6632 0
2 0.6066 0
2 -0.70758 0
2 4.4814 3.8164
As you can see, sequentialfs has generated a much more parsimonious model than the ordinary least squares regression. We also have a model that should generate significantly more accurate predictions. Moreover, if we needed to (hypothetically) extract the five most important variables we could simple grab the output from Steps 1:5.
Run a Simulation
As a last step, we'll once again run a simulation to benchmark the technique. Here, once again, we can see that the feature selection technique is doing a very good job identifying the true predictors while screening out the distractors.
matlabpool open sumin = 0; parfor j=1:100 Z12 = randn(100,40); Z1 = randn(100,1); X = Z12 + repmat(Z1, 1,40); B0 = zeros(10,1); B1 = ones(10,1); Beta = 2 * vertcat(B0,B1,B0,B1); Y = X * Beta + 15*randn(100,1); fun = @(x0,y0,x1,y1) norm(y1-x1*(x0\y0))^2; % residual sum of squares [in,history] = sequentialfs(fun,X,Y,'cv',5); sumin = in + sumin; end bar(sumin) matlabpool close
Starting matlabpool using the 'local4' configuration ... connected to 3 labs. Sending a stop signal to all the labs ... stopped.

One last comment: As you can see, neither sequentialfs nor all possible subset selection is perfect. Neither of these techniques was able to capture all of the true variables while excluding the distractors. It's important to recognize that these data sets were deliberately designed to be difficult to fit. [We don't want to benchmark performance using "easy" problems where any old technique might succeed.]
Next week's blog post will focus on a radically different way to solve these same types of problems. Stay tuned for a look at regularization techniques such as ridge regression, lasso and the elastic net.
Here are a couple questions that you might want to consider: # All possible subset methods are O(2^n). What is the order of complexity of sequential feature selection? # Other than scaling issues, can you think of any possible problems with sequential feature selection? Are there situations where the algorithm might miss important variables?
- Category:
- Curve fitting,
- Statistics







Comments
To leave a comment, please click here to sign in to your MathWorks Account or create a new one.