Minimizing energy to segment images, or cluster data
Brett‘s Pick this week is Pottslab – Multilabel segmentation of vectorial data, by Martin.
Segmenting images and clustering data are very common challenges, for which MATLAB (and MathWorks Toolboxes) provide numerous solutions. Martin’s Potts-energy-based approach works beautifully for some difficult cases. Consider the ‘lighthouse.png’ image that ships with the Image Processing Toolbox:

If we wanted to “cluster” the colors of this images, we could:
- Simply specify the number of clusters, and use rgb2ind():
nClusters = 9; [X,map] = rgb2ind(img,nClusters,'nodither'); imshow(X,map)

- Actually do some statistical (e.g., k-means) clustering:
rc = img(:,:,1); gc = img(:,:,2); bc = img(:,:,3); segcolors = double([rc(:) gc(:) bc(:)]); Idx = kmeans(segcolors,nClusters,... 'distance','sqEuclidean',... 'start',map,... 'emptyaction','singleton'); clusteredImg = reshape(Idx,size(rc)); imshow(clusteredImg,map)

- Use an energy-minimation approach like the one Martin provided:
img = imresize(img, 0.5);
gamma = 0.15;
PottsImage = minL2Potts2DADMM(img, gamma, 'verbose', true);
energyU = energyL2Potts(PottsImage, img, gamma);
energyImg = energyL2Potts(img, img, gamma);
imshow(PottsImage)
title(sprintf('Potts segmentation (Potts energy: %.1f)', energyU));

In some regards, the Potts-energy approach provides superior results. And I didn’t have to specify the number of cluster a priori . Some observations, though:
- I didn’t show it, but the “Potts energy” of the original (resized) image was about an order of magnitude higher than the energy of the clustered image.
- You’ll notice the call to imresize before I called the Potts functionality. This method is extremely memory intensive, and runs out of memory on my computer when I try to operate on the full-size original image, even though it’s pretty small (640 x 480 x 3). (You might have good success upsampling [with imresize] the PottsImage to align the clustering [and segmentations based thereon] to the original image. But this can also introduce “noise,” and extra computational expense, to the problem.)
- The Potts approach is relatively very slow, even on the smaller images. The rgb2ind computation took 17 msec on my laptop. The kmeans approach took 1.2 seconds. The Potts-energy approach took 3.3 seconds, including the necessary resizing.
Still, options are always good, and I’m very impressed with the quality of the Potts clustering. (Notice the quality of the roof segmentation, for instance.)
Martin’s code is quite extensive, well-implemented (in object-oriented code), and documented. It is also much more flexible than I’ve demonstrated here. My focus is typically on image analysis, but the Potts-energy minimization can be applied to signal based operations, too, like denoising:
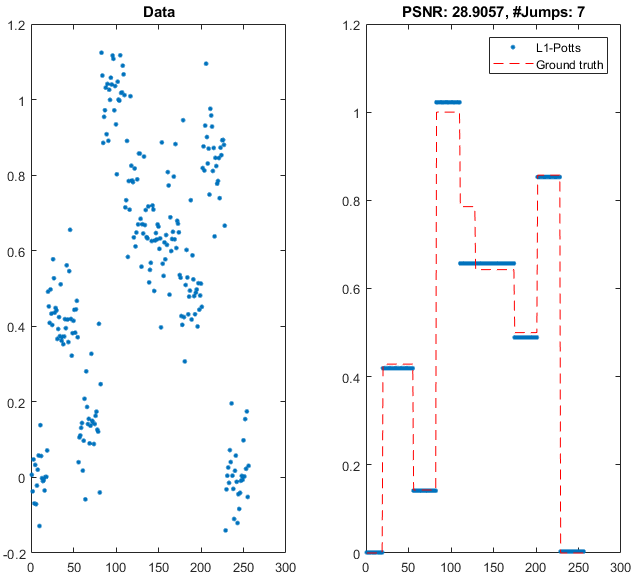
Thanks, Martin, for sharing this very interesting and useful functionality!
As always, I welcome your thoughts and comments.
Published with MATLAB® R2017b
- Category:
- Picks
 Cleve’s Corner: Cleve Moler on Mathematics and Computing
Cleve’s Corner: Cleve Moler on Mathematics and Computing The MATLAB Blog
The MATLAB Blog Guy on Simulink
Guy on Simulink MATLAB Community
MATLAB Community Artificial Intelligence
Artificial Intelligence Developer Zone
Developer Zone Stuart’s MATLAB Videos
Stuart’s MATLAB Videos Behind the Headlines
Behind the Headlines File Exchange Pick of the Week
File Exchange Pick of the Week Hans on IoT
Hans on IoT Student Lounge
Student Lounge MATLAB ユーザーコミュニティー
MATLAB ユーザーコミュニティー Startups, Accelerators, & Entrepreneurs
Startups, Accelerators, & Entrepreneurs Autonomous Systems
Autonomous Systems Quantitative Finance
Quantitative Finance MATLAB Graphics and App Building
MATLAB Graphics and App Building








Comments
To leave a comment, please click here to sign in to your MathWorks Account or create a new one.