Reading the last N lines from a CSV file
Jiro‘s pick this week is csvreadtail by Mike.
Recently, I’ve been working on customer projects around improving the performance of analysis code. These projects typically involve using the Profiler to find bottlenecks, vectorizing code wherever possible, and using the appropriate data types that fit the particular tasks. Regarding the last point, I was pleasantly surprised to find out how string manipulations were so much more efficient using the new string data type rather than character arrays or cell array of chars.
In some cases, the bottlenecks I found with the Profiler could be solved by vectorizing or changing the algorithm. But one area that I found a bit challenging was when the bottleneck involved file I/O. When you need to read or write from/to files, there’s not much you can do in terms of speeding up that process.
Well, that’s not entirely true. With readtable or datastore, you can specify import options to optimize how you import your data. You can do something similar with textscan by specifying a format spec.
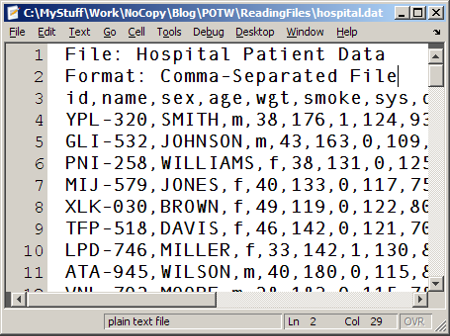
My pick this week falls into this category of efficient file reading. The use case is quite specific, but say you just need to read in the last N lines from a large CSV file. If you know beforehand how many lines the files has, csvread has an option for specifying the range. If you don’t know the number of lines, how can you read the last N lines? Would you need to first scan the file to figure out how many lines it has? Mike’s csvreadtail uses a clever technique of moving the file position, using fseek, to the end of the file and then “reading back” until a specified number of lines have been read. Great idea!

I do have a few suggestions for improvement, though. I’m pretty certain these will make this code much more efficient.
- This one is not about efficiency, but in the parsecsv.m helper function, strsplit is used to split up a line of text (with commas) into a vector. That command should set the CollapseDelimiters options to false, in order to accurately capture empty entries.
strsplit(s{iLine},',','CollapseDelimiters',false)- Currently, the way the file position is moved back is by repeatedly calling fseek relative to the end of file. This can be done more efficiently by moving relative to the current position. Since the algorithm reads one byte at a time, you can move back 2 bytes from the current position.
- Rather than reading one byte at a time, it is more efficient to read a chunk of data (several hundreds or thousands of bytes) at a time. The tricky part is figuring out how many bytes to go back and read. Perhaps one approach is to adaptively change the amount to go back. For example, depending on how many lines of data were read with a single read, you can read more (or less) next time.
Tall Arrays
For those of you on R2016b or newer, you may be interested in checking out the new tall array capability. Tall arrays allow you to work with large data sets in out-of-memory fashion, only loading parts of data necessary for analysis. In that framework, the tail function reads in the last N rows of data.
Comments
Give it a try and let us know what you think here or leave a comment for Mike.
- Category:
- Picks
 Cleve’s Corner: Cleve Moler on Mathematics and Computing
Cleve’s Corner: Cleve Moler on Mathematics and Computing The MATLAB Blog
The MATLAB Blog Guy on Simulink
Guy on Simulink MATLAB Community
MATLAB Community Artificial Intelligence
Artificial Intelligence Developer Zone
Developer Zone Stuart’s MATLAB Videos
Stuart’s MATLAB Videos Behind the Headlines
Behind the Headlines File Exchange Pick of the Week
File Exchange Pick of the Week Hans on IoT
Hans on IoT Student Lounge
Student Lounge MATLAB ユーザーコミュニティー
MATLAB ユーザーコミュニティー Startups, Accelerators, & Entrepreneurs
Startups, Accelerators, & Entrepreneurs Autonomous Systems
Autonomous Systems Quantitative Finance
Quantitative Finance MATLAB Graphics and App Building
MATLAB Graphics and App Building





Comments
To leave a comment, please click here to sign in to your MathWorks Account or create a new one.